Dataset I - Building a CME source region catalogue
The purpose of this code journey is to document the process behind the building of both the source region catalogue . The idea is that this should serve as a reference to ensure that the building process is transparent and to prevent confusion about what decisions were applied, when and why.Although closely related, there are two independent tasks here with a sequential dependence as well as some pre-processing that needs to be done:
-
Pre-processing: Ensuring that the SHARPs bounding boxes are clean. This includes
a. Removing SHARPs that are too large: Some entries seem to be "glitchy", with bounding boxes occupying almost the whole solar surface. Before anything is done, we need to deal with this.
b. Trimming bounding boxes: Some bounding boxes (for large regions) extend beyond the limb at the start and end of the region's lifetime. These bounding boxes need to be trimmed. However, this shouldn't affect calculations of the average area of the bounding box.
c. Removing overlapping bounding boxes: Unfortunately, there are many cases where either a bounding box is contained whole inside another one or a great part (%) is contained inside another one. This makes difficult associating events to a particular region, specially if it falls within the overlap. But even if it falls outside the overlap, it reduces the confidence we can have on that association. Thus, overlaps must be dealt with.
-
Associating CMEs to active regions: Using the series of scripts already developed. They should make use of the processed bounding boxes and results of these script will be stored in the same database.
-
Building a general dataset using sequences: Described TBD
-
Preparing the previous dataset to be used with SDOML: TBD.
In the following I will detail the steps taken to achieve what is described in each of the steps above.
Pre-processing
Data loading
Creating database
The first step is to create a database that will hold all the information needed. The schema for the database is as follows
CAUTION: The image_timestamp columns
While the name of these columns may hint at them referring to the timestamp of a solar image, in reality this is remnant from when I was considering this to be limited to SDOML images. However, since them I've split the processes and all here should be independent of SDOML. What this means is that image_timestamps here will refer simply to SHARPs-like timestamp (that is, with a 00, 12, 24, 36 or 48 minutes part).
CREATE TABLE HARPS (
harpnum INTEGER PRIMARY KEY, -- Unique identifier for each HARP region
start TEXT NOT NULL, -- Start timestamp of the HARP region
end TEXT NOT NULL, -- End timestamp of the HARP region
pix_width INTEGER, -- Refers to the fixed width in pixel values used for cutouts
pix_height INTEGER, -- Refers to the fixed height in pixel values used for cutouts
area FLOAT, -- Refers to the area of the HARP region
CHECK (area >= 0),
CHECK (pix_width >= 0),
CHECK (pix_height >= 0),
CHECK (end >= start)
);
CREATE TABLE CMES (
cme_id INTEGER NOT NULL PRIMARY KEY, -- Unique identifier for each CME
cme_date TEXT NOT NULL, -- Date and time of the CME
cme_pa REAL, -- Position angle of the CME
cme_width REAL NOT NULL, -- Width of the CME
cme_halo INTEGER, -- Indicator for Halo CMEs (1 for Halo, else NULL)
cme_seen_in INTEGER NOT NULL, -- Where the CME was observed (e.g., C2, C3)
cme_three_points INTEGER NOT NULL, -- Number of observed points for the CME
cme_quality INTEGER NOT NULL, -- Quality rating for the CME observation
image_timestamp TEXT
CHECK (
((cme_pa IS NULL) = (cme_halo = 1)) -- Ensure Halo CMEs don't have a position angle
)
);
CREATE TABLE FLARES (
flare_id INTEGER NOT NULL PRIMARY KEY, -- Unique identifier for each flare
harpnum INTEGER REFERENCES HARPS (harpnum), -- Associated HARP region
flare_date TEXT NOT NULL, -- Date and time of the flare
flare_lon REAL, -- Longitude of the flare
flare_lat REAL, -- Latitude of the flare
flare_class_score REAL NOT NULL, -- Score indicating the class of the flare
flare_class TEXT NOT NULL, -- Class of the flare (e.g., M1, X2)
flare_ar INTEGER, -- Active Region number of the flare
flare_ar_source TEXT, -- Source of the Active Region number
flare_verification TEXT, -- Verification status of the flare information
image_timestamp TEXT REFERENCES
CHECK (
(flare_ar IS NULL AND flare_lon IS NOT NULL AND flare_lat IS NOT NULL) OR
(flare_ar IS NOT NULL) -- Ensure either flare_ar or both flare_lon and flare_lat are provided
)
);
CREATE TABLE RAW_HARPS_BBOX (
-- Reference to the harpnum column in the harps table
harpnum INTEGER REFERENCES HARPS (harpnum),
-- Reference to the timestamp column in the images table
timestamp TEXT,
-- Minimum longitude of BBOX
LONDTMIN REAL,
-- Maximum longitude of BBOX
LONDTMAX REAL,
-- Minimum latitude of BBOX
LATDTMIN REAL,
-- Maximum latitude of BBOX
LATDTMAX REAL,
-- Is Rotated Bounding Box? Specifies if the BBOX is calculated using differential solar rotation because it was missing from the SHARP dataset
IRBB INTEGER,
-- Is trusted magnetic field data?
IS_TMFI INTEGER,
PRIMARY KEY (harpnum, timestamp),
-- Need to check that the timestamp has minutes 12, 24, 36, 48, or 00
CHECK (strftime('%M', timestamp) IN ('12', '24', '36', '48', '00'))
);
CREATE TABLE PROCESSED_HARPS_PIXEL_BBOX (
-- Reference to the harpnum column in the harps table
harpnum INTEGER REFERENCES HARPS (harpnum),
-- Reference to the timestamp column in the images table
timestamp TEXT,
-- Minimum x-coordinate of the bounding box; value must be between 0 and 511
x_min INTEGER NOT NULL CHECK (x_min >= 0 AND x_min <= 511),
-- Maximum x-coordinate of the bounding box; value must be between 0 and 511
x_max INTEGER NOT NULL CHECK (x_max >= 0 AND x_max <= 511),
-- Minimum y-coordinate of the bounding box; value must be between 0 and 511
y_min INTEGER NOT NULL CHECK (y_min >= 0 AND y_min <= 511),
-- Maximum y-coordinate of the bounding box; value must be between 0 and 511
y_max INTEGER NOT NULL CHECK (y_max >= 0 AND y_max <= 511),
-- x-coordinate of the center of the bounding box; value must be between 0 and 511
x_cen INTEGER NOT NULL CHECK (x_cen >= 0 AND x_cen <= 511),
-- y-coordinate of the center of the bounding box; value must be between 0 and 511
y_cen INTEGER NOT NULL CHECK (y_cen >= 0 AND y_cen <= 511), overlap_percent REAL, overlap_harpnum INTEGER, area REAL,
-- Unique key for the table based on harpnum and timestamp
PRIMARY KEY (harpnum, timestamp),
-- References the harps_bbox table to maintain a relationship with HARPS bounding box information
FOREIGN KEY (harpnum, timestamp) REFERENCES HARPS_BBOX (harpnum, timestamp)
);
CREATE TABLE FINAL_CME_HARP_ASSOCIATIONS (
cme_id INTEGER UNIQUE NOT NULL REFERENCES CMES(cme_id), -- Unique identifier for each CME
harpnum INTEGER NOT NULL REFERENCES HARPS(harpnum), -- Associated HARP region
association_method TEXT NOT NULL, -- Method used to determine the association
verification_score REAL, -- Confidence score of the association
independent_verified INTEGER NOT NULL DEFAULT 0, -- Verification level for association (0 if only by me, 1 if verified by external)
PRIMARY KEY (cme_id, harpnum) -- Unique pairing of CME and HARP
);
CREATE TABLE BBOX_OVERLAPS (
harpnum_a INTEGER REFERENCES HARPS(harpnum),
harpnum_b INTEGER REFERENCES HARPS(harpnum),
mean_overlap REAL,
mean_pixel_overlap REAL,
ocurrence_percentage REAL,
pixel_ocurrence_percentage REAL,
harpnum_a_area REAL,
harpnum_a_pixel_area REAL,
harpnum_b_area REAL,
coexistence REAL
);
CREATE TABLE OVERLAP_RECORDS (
harpnum_a INTEGER REFERENCES HARPS(harpnum),
harpnum_b INTEGER REFERENCES HARPS(harpnum),
decision STRING,
mean_overlap REAL,
std_overlap REAL,
ocurrence_percentage REAL,
harpnum_a_area REAL,
harpnum_b_area REAL,
b_over_a_area_ratio REAL,
PRIMARY KEY (harpnum_a, harpnum_b),
CHECK (harpnum_a_area < harpnum_b_area),
CHECK (decision IN ('MERGED A WITH B', 'DELETED A IN FAVOR OF B'))
);
CREATE TABLE PROCESSED_HARPS_BBOX(
-- Reference to the harpnum column in the harps table
harpnum INTEGER REFERENCES HARPS (harpnum),
-- Reference to the timestamp column in the images table
timestamp TEXT,
-- Minimum longitude of BBOX
LONDTMIN REAL,
-- Maximum longitude of BBOX
LONDTMAX REAL,
-- Minimum latitude of BBOX
LATDTMIN REAL,
-- Maximum latitude of BBOX
LATDTMAX REAL,
-- Is Rotated Bounding Box? Specifies if the BBOX is calculated using differential solar rotation because it was missing from the SHARP dataset
IRBB INTEGER,
-- Is trusted magnetic field data?
IS_TMFI INTEGER,
PRIMARY KEY (harpnum, timestamp),
-- Need to check that the timestamp has minutes 12, 24, 36, 48, or 00
CHECK (strftime('%M', timestamp) IN ('12', '24', '36', '48', '00'))
);
CREATE TABLE MAJUMDAR_SRC (
cme_id INTEGER PRIMARY KEY REFERENCES CMES (cme_id),
cme_date TEXT NOT NULL,
cme_pa REAL,
cme_width REAL,
cme_speed REAL,
sr_type TEXT,
sr_lat REAL NOT NULL,
sr_lon REAL NOT NULL
, matching_harps INTEGER REFERENCES HARPS (harpnum) DEFAULT NULL, dist REAL, sr_pa REAL);
CREATE TABLE IF NOT EXISTS CMES_HARPS_EVENTS (
harpnum INTEGER,
cme_id INTEGER,
flare_id INTEGER REFERENCES FLARES(flare_id),
flare_hours_diff INTEGER NOT NULL,
dimming_id INTEGER REFERENCES DIMMINGS(dimming_id),
dimming_hours_diff INTEGER NOT NULL,
FOREIGN KEY(harpnum,cme_id) REFERENCES CMES_HARPS_SPATIALLY_CONSIST(harpnum,cme_id),
PRIMARY KEY(harpnum,cme_id)
);
CREATE TABLE IF NOT EXISTS CMES_HARPS_SPATIALLY_CONSIST (
harpnum INTEGER NOT NULL REFERENCES HARPS(harpnum),
cme_id INTEGER NOT NULL REFERENCES CMES(cme_id),
PRIMARY KEY(harpnum,cme_id)
);
CREATE TABLE IF NOT EXISTS DIMMINGS (
dimming_id INTEGER NOT NULL,
harpnum INTEGER REFERENCES HARPS(harpnum),
harps_dimming_dist REAL NOT NULL,
dimming_start_date TEXT NOT NULL,
dimming_peak_date TEXT NOT NULL,
dimming_lon REAL NOT NULL,
dimming_lat REAL NOT NULL,
image_timestamp TEXT REFERENCES
PRIMARY KEY(dimming_id)
);
CREATE TABLE NOAA_HARPNUM_MAPPING (
noaa INTEGER,
harpnum INTEGER REFERENCES HARPS (harpnum),
PRIMARY KEY (noaa, harpnum)
);
CREATE TABLE NOAAS (
noaa INTEGER PRIMARY KEY
);
CREATE TABLE SANJIV_SRC (
cme_id INTEGER PRIMARY KEY REFERENCES CMES (cme_id),
cme_date TEXT,
cme_width REAL,
cme_speed REAL,
flare_time TEXT,
flare_class TEXT,
noaa INTEGER REFERENCES NOAAS (noaa)
);
CREATE TABLE HOURLY_BBOX(
-- Reference to the harpnum column in the harps table
harpnum INTEGER REFERENCES HARPS (harpnum),
-- Reference to the timestamp column in the images table
timestamp TEXT,
-- Minimum longitude of BBOX
LONDTMIN REAL,
-- Maximum longitude of BBOX
LONDTMAX REAL,
-- Minimum latitude of BBOX
LATDTMIN REAL,
-- Maximum latitude of BBOX
LATDTMAX REAL,
-- Is Rotated Bounding Box? Specifies if the BBOX is calculated using differential solar rotation because it was missing from the SHARP dataset
IRBB INTEGER,
-- Is trusted magnetic field data?
IS_TMFI INTEGER,
PRIMARY KEY (harpnum, timestamp),
-- Need to check that the timestamp has minutes 12, 24, 36, 48, or 00
CHECK (strftime('%M', timestamp) IN ('12', '24', '36', '48', '00'))
);
CREATE TABLE HOURLY_PIXEL_BBOX(
-- Reference to the harpnum column in the harps table
harpnum INTEGER REFERENCES HARPS (harpnum),
-- Reference to the timestamp column in the images table
timestamp TEXT,
-- Minimum x-coordinate of the bounding box; value must be between 0 and 511
x_min INTEGER NOT NULL CHECK (x_min >= 0 AND x_min <= 511),
-- Maximum x-coordinate of the bounding box; value must be between 0 and 511
x_max INTEGER NOT NULL CHECK (x_max >= 0 AND x_max <= 511),
-- Minimum y-coordinate of the bounding box; value must be between 0 and 511
y_min INTEGER NOT NULL CHECK (y_min >= 0 AND y_min <= 511),
-- Maximum y-coordinate of the bounding box; value must be between 0 and 511
y_max INTEGER NOT NULL CHECK (y_max >= 0 AND y_max <= 511),
-- x-coordinate of the center of the bounding box; value must be between 0 and 511
x_cen INTEGER NOT NULL CHECK (x_cen >= 0 AND x_cen <= 511),
-- y-coordinate of the center of the bounding box; value must be between 0 and 511
y_cen INTEGER NOT NULL CHECK (y_cen >= 0 AND y_cen <= 511), overlap_percent REAL, overlap_harpnum INTEGER, area REAL,
-- Unique key for the table based on harpnum and timestamp
PRIMARY KEY (harpnum, timestamp),
-- References the harps_bbox table to maintain a relationship with HARPS bounding box information
FOREIGN KEY (harpnum, timestamp) REFERENCES HARPS_BBOX (harpnum, timestamp)
);Loading SWAN-SF bounding boxes
In this database we load the SWAN-SF data (bounding boxes). While the original, raw data contains the LON_MIN, LON_MAX, LAT_MIN, LAT_MAX keywords, these refer to the boundaries of the patch of the AR that was used to calculate the magnetic field parameters. This is a highly variable bounding box, and for some timestamps is non-existent (resulting in all keywords being NaN). Instead of this we're interested in the LONDTMIN, LONDTMAX, LATDTMIN, LATDTMAX keywords which correspond to a bounding box that doesn't change over time (width, height are constant) and so it tracks the AR more consistently for the purposes of event assignment and image cutout extraction. As mentioned before, these keywords are not included in SWAN-SF and for some timestamps all keywords are NaN. Therefore we
- Extract keywords
LONDTMIN, LONDTMAX, LATDTMIN, LATDTMAXfrom the JSOC. - Fill missing values by using sunpy's
propagate_with_solar_surface()which calculates new coordinates based on a differential rotation model.
These values are then loaded into the HARPS_BBOX table. Based on the first and last timestamp available for each individual harpnum value in the table, we create a new entry in the HARPS table with these values as start and end respectively. This is because no subsequent steps should modify (remove) any timestamp and so assigning the first and last values at this step should be safe.
Loading the LASCO CME catalogue
The lasco CME catalogue can be found in .txt format here. We parse this catalogue, including the comments. We add the following columns
-
cme_quality: 0 if no comments, 1 if Poor and 2 if Very Poor. -
cme_seen_in: 0 if seen in both C2 and C3, 1 if seen only in C2 and 2 if seen only in C3. -
cme_three_points: When there's a warning about there only beingnpoints for calculations about the CME we record the value ofnhere. -
cme_halo: 1 if the cme is halo and 0 otherwise.
Apart from this, we keep the same columns, although we make the cme_pa value NULL for halo cmes. This data is loaded into the CMES table with the ID of each CME formed by the date in format YYYYMMDDHHMMSS followed by the position angle of the CME formed of three digits (e.g. for 89 we use 089) and with 999 for halo CMEs. For example, one index could be 20231923142316034. This ensures every ID is unique.
Aside: Repeated IDs
Although these IDs should never be identical between two CMEs, we surprisingly find that somehow the catalogue has recorded two instances were two CMEs with the exact same timestamp and exact same position angle but different width are recorded. How this happened is a mystery, but given it occurs only with 4 CMEs in total and that it doesn't seem plausible that two CMEs with the same timestamp and position angle but different width exist, we simply remove the duplicate entries. The IDs of the CMEs that are removed are 20060517230604238, 20110918112407076
The data is then loaded using
df = pd.read_csv(LASCO_CME_DATABASE)
no_duplicates = df.drop_duplicates(subset="CME_ID", keep=False)
for i, row in no_duplicates.drop_duplicates(subset=["CME_ID"], keep="first").iterrows():
new_conn.execute(
"""
INSERT INTO CMES (cme_id, cme_date, cme_pa, cme_width, cme_halo, cme_seen_in, cme_quality, cme_three_points)
VALUES (?, ?, ?, ?, ?, ?, ?, ?)
""",
(
int(row["CME_ID"][2:]),
row["CME_DATE"],
row["CME_PA"],
row["CME_WIDTH"],
row["CME_HALO"],
row["CME_SEEN_IN"],
row["CME_QUALITY"],
row["CME_THREE_POINTS"]
)
)Calculating areas
The areas of the SHARPs regions is a relevant quantity in future steps (i.e. the calculation of overlaps). Therefore, we calculate the area of each harpnum by calculating the area at each timestamp and averaging all the areas (all areas should be almost the same). To calculate the area we use
WITH AREAS AS (
SELECT harpnum,
COALESCE(AVG(NULLIF(100.0 * ((PI()/180.0 * (LONDTMAX - LONDTMIN) * ABS(SIN(PI()/180.0 * LATDTMAX) - SIN(PI()/180.0 * LATDTMIN))) / (2.0 * PI())), 0)),0) AS area
FROM RAW_HARPS_BBOX
GROUP BY harpnum
)
UPDATE HARPS
SET area = (SELECT area FROM AREAS WHERE AREAS.harpnum = HARPS.harpnum);which makes use of the following equation to calculate the spherical surface area of a region
But divided by to get the percentage of the visible solar surface that the region occupies. The NULLIF function is used to replace area values of 0 with a NULL so that values that incorrectly could result in area 0 do not count towards the AVG. The COALESCE simply replaces an average of NULL with 0, in case there are no valid values for a particular harpnum.
With these values we can see what the distribution of areas looks like
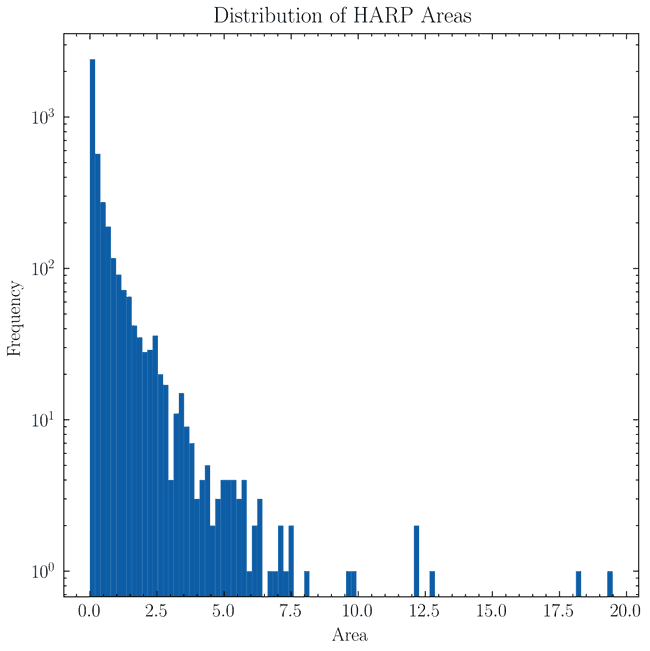
and so it's clear hat almost all regions have areas % of the visible solar surface. However, there are two regions with areas larger than % of the solar surface. These are 6713 and 4225 which have been verified manually to be glitchy bounding boxes. Therefore, we remove these two regions from the database.
Now, while in this way we have dealt with glitchy regions, there remains a more complex problem to tackle. While most sharps are relatively well-sized, some of them occupy a good fraction of the visible solar surface when compared to others. If we look at the distribution of the number of NOAA ARs contained in each SHARPs region
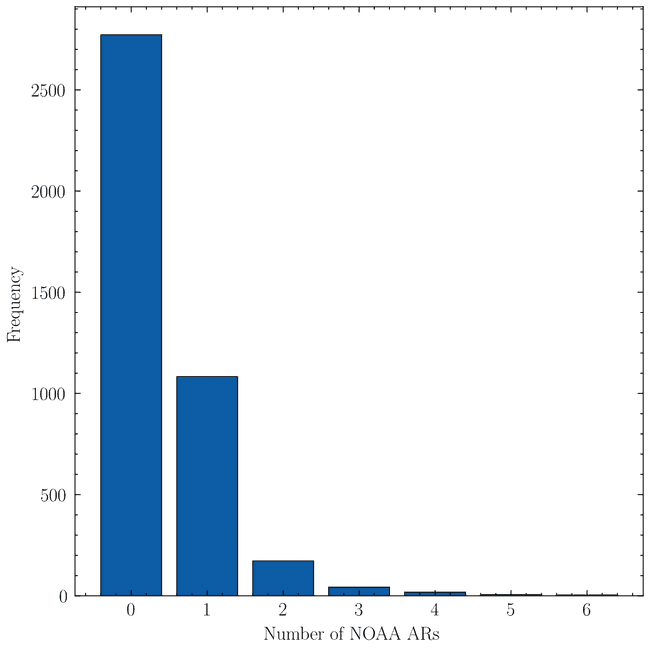
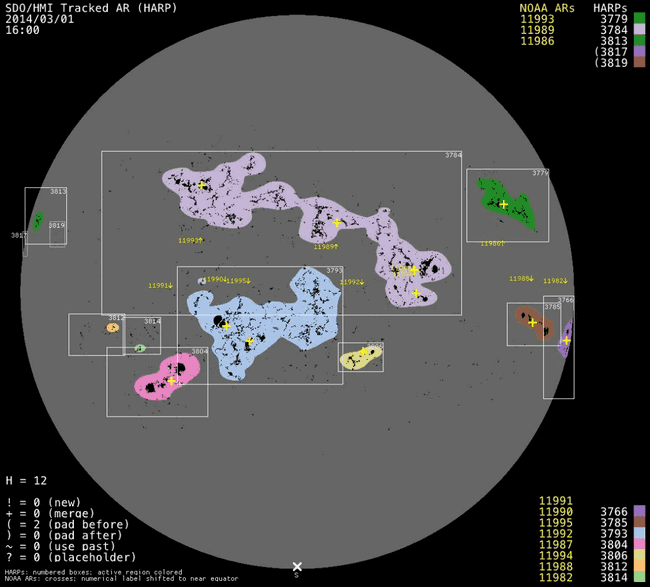
we find surprisingly that most SHARPs don't have any corresponding NOAA AR. We are aware thanks to work in SWAN-SF that some of these associations are missing or are wrong. However, these should affect at most about ~100 SHARPs regions. A good number of regions also have single NOAA AR. However, where the real problem lies is where regions have a large number (say e.g. four of more) NOAA ARs inside them. Although rare, we do not want to dismiss these as simply large SHARP regions. Ultimately, if many different magnetic regions are included under the same SHARP region, then the SHARP region is not a good representation of any of them. See for example
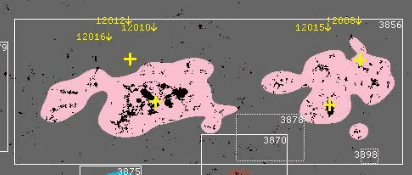
where we can see that the SHARP region contains 4 different magnetic regions, yet it could probably be split into two. However, looking at AIA 193 images
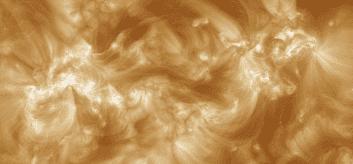
It may be that grouping them together is correct. In fact, looking at another region (3784)
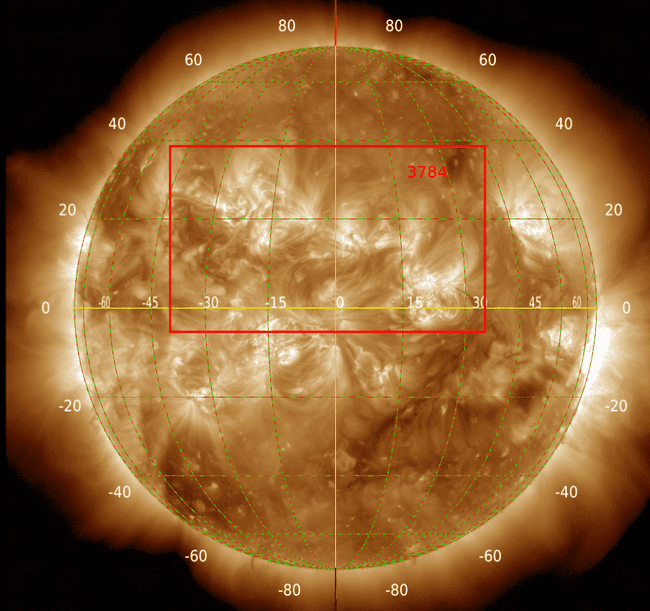
it also seems to be right. In any case, these big regions pose a serious problem as they may overlap a lot with other regions and can be more prone to missasociating events that didn't originate in them, yet at the same time they may be the most active regions. For the moment, they are kept in the database but I will say the following:

where we can see that the SHARP region contains 4 different magnetic regions, yet it could probably be split into two. However, looking at AIA 193 images

It may be that grouping them together is correct. In fact, looking at another region (3784)


it also seems to be right. In any case, these big regions pose a serious problem as they may overlap a lot with other regions and can be more prone to missasociating events that didn't originate in them, yet at the same time they may be the most active regions. For the moment, they are kept in the database but I will say the following:
-
Removing them: On the one hand it can help with the problem of overlaps. However, in order to be consistent, when removing them we should remove all timestamps during the lifetime of that region meaning we lose every region in the Sun potentially for several days. Why I say that we should remove everything is because otherwise dimmings that do belong to this removed region could be assigned to a close but false region after removing it.
This would still not solve problems at the edge of the intervals (really only at the ending edge) where a dimming may also be associated with another false region if it's close enough in time and space.
-
Keeping them: Doesn't create all the headaches mentioned above. However, it may be that the region is so large that it overlaps with many others, making me drop them from the catalogue based on rules defined later. It could also be that the region being so large I associate events that have nothing to do with it (dimmings, since flares are verified independently).
All in all, it seems best to leave them and just assess how many of them show up in the final associations.
So in conclusion, given the areas I drop the two glitchy regions and keep the rest. These are dropped by doing
CREATE TEMPORARY TABLE NO_BIG_HARPS AS
SELECT RHB.* FROM RAW_HARPS_BBOX RHB
INNER JOIN HARPS H ON RHB.harpnum = H.harpnum
WHERE H.area < 18Trimming bounding boxes
As mentioned before, some bounding boxes extend beyond the limb of the Sun, see for example
| harpnum | timestamp | LONDTMIN | LONDTMAX |
|---|---|---|---|
| 4225 | 2014-06-09 16:48:00 | -144 | -81 |
| 4225 | 2014-06-12 01:12:00 | -113 | -49 |
| 4225 | 2014-06-23 14:48:00 | 41 | 104 |
| 4225 | 2014-06-23 12:24:00 | 39 | 103 |
| 4225 | 2014-06-13 13:48:00 | -93 | -29 |
and even in some cases the whole bounding box is outside the limb. So we do the following
CREATE TEMPORARY TABLE NO_BIG_HARPS AS
SELECT RHB.* FROM RAW_HARPS_BBOX RHB
INNER JOIN HARPS H ON RHB.harpnum = H.harpnum
WHERE H.area < 18;
CREATE TEMPORARY TABLE TRIMMED_HARPS_BBOX AS
SELECT NBH.* FROM NO_BIG_HARPS NBH-
For bounding boxes completely behind the limb: We remove them from the database.
DELETE FROM TRIMMED_HARPS_BBOX WHERE (LONDTMIN < -90 AND LONDTMAX < -90) OR (LONDTMIN > 90 AND LONDTMAX > 90) -
For bounding boxes where one edge extends beyond the limb: We trim that edge to be either (left side) or right side.
UPDATE TRIMMED_HARPS_BBOX SET LONDTMIN = -90 WHERE LONDTMIN < -90; UPDATE TRIMMED_HARPS_BBOX SET LONDTMAX = 90 WHERE LONDTMAX > 90;
Calculating overlaps
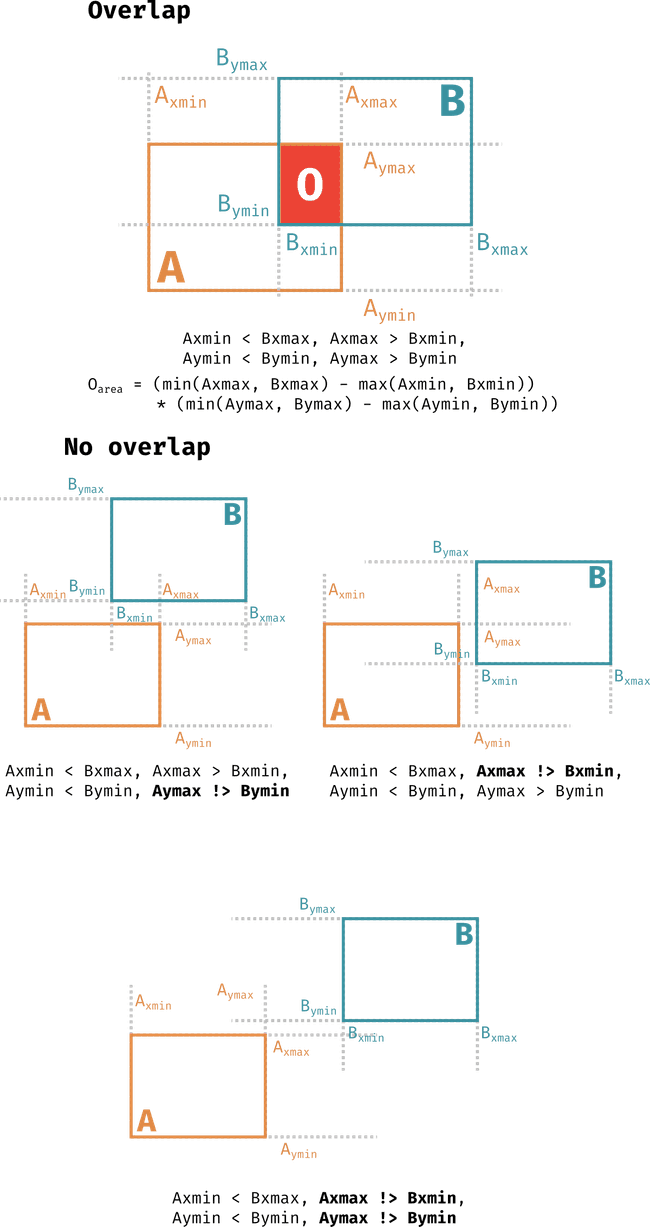
The figure above details the rules for when there's an overlap between two bounding boxes and also how these rules are broken when there are no overlaps. When these conditions are met one may calculate the area of the overlap can be calculated according to the equation for in cartesian coords and similarly for spherical areas by replacing with longitudes, with latitudes and adding the relevant . Following this, we calculate the overlap between each HARPs region and every other HARPs region present at the Sun at the same timestamp for all timestamps. This is done as follows
CREATE TEMPORARY TABLE temp_overlap AS
SELECT
a.harpnum AS harpnum1,
b.harpnum AS harpnum2,
a.timestamp AS timestamp,
100.0 * CASE WHEN a.LONDTMIN < b.LONDTMAX AND a.LONDTMAX > b.LONDTMIN AND a.LATDTMIN < b.LATDTMAX AND a.LATDTMAX > b.LATDTMIN
THEN (MIN(a.LONDTMAX, b.LONDTMAX) - MAX(a.LONDTMIN, b.LONDTMIN)) * ABS((SIN(PI() / 180.0 * MIN(a.LATDTMAX, b.LATDTMAX)) - SIN(PI() / 180.0 * MAX(a.LATDTMIN, b.LATDTMIN))))
ELSE 0
END / ((2.0 * PI() / (100.0 * PI() / 180.0)) * H.area) AS overlap_percent
FROM TRIMMED_HARPS_BBOX a
JOIN TRIMMED_HARPS_BBOX b ON a.timestamp = b.timestamp AND a.harpnum != b.harpnum
JOIN HARPS H ON H.harpnum = a.harpnum;Notice we divide the overlap by the area of the first region. The constant multiplying H.area is to remove the constants we have included in the area calculation (dividing by and multiplying by 100 as well as converting the longitude to radians as these would cancel out in the division). The result here is a table containing harpnum1, harpnum2 combinations for every timestamp they coexist with their overlap calculation. While this is the most complete representation of the overlaps between regions, it is not the most useful. Instead, we want to know the average overlap between two regions over their whole lifetime so that we may evaluate if they are too similar. To do this we do
SELECT tpo.harpnum1,
tpo.harpnum2,
AVG(tpo.overlap_percent) AS mean_overlap,
(100.0 * (strftime('%s', MAX(tpo.timestamp)) - strftime('%s', MIN(tpo.timestamp)))) / (1.0 * NULLIF(strftime('%s', H.end) - strftime('%s', H.start), 0)) AS ocurrence_percentage
FROM temp_overlap tpo
JOIN HARPS H ON H.harpnum = tpo.harpnum1
WHERE tpo.overlap_percent > 0
GROUP BY harpnum1, harpnum2;In this operation, we also calculate the percentage of harpnum1's lifetime that the overlap between the two regions occurred. However, note that how we do this is by assuming that there are no missing timestamps between the first and last timestamp where they overlap and that the regions don't overlap, stop overlapping and then overlap again. An alternative way is to count how many rows there are, multiply by 12 minutes and then use that as the overlap duration.
On closer inspection, turns out there's a difference although small
| harpnum1 | harpnum2 | mean_overlap | ocurrence_percentage | alternative_ocurrence_percentage | diff |
|---|---|---|---|---|---|
| 537 | 542 | 0.80 | 75.64 | 74.03 | 1.61 |
| 542 | 537 | 3.27 | 100.00 | 97.87 | 2.13 |
| 1527 | 1537 | 0.03 | 12.99 | 12.33 | 0.66 |
| 1537 | 1527 | 1.55 | 63.14 | 59.94 | 3.21 |
| 1694 | 1698 | 0.40 | 57.14 | 44.33 | 12.81 |
| 1698 | 1694 | 0.39 | 71.60 | 55.56 | 16.05 |
| 1967 | 1998 | 0.01 | 1.51 | 1.16 | 0.35 |
| 1998 | 1967 | 0.02 | 2.54 | 1.96 | 0.59 |
| 4359 | 4360 | 0.02 | 3.61 | 3.51 | 0.10 |
| 4360 | 4359 | 0.03 | 5.72 | 5.56 | 0.16 |
| 6115 | 6131 | 0.14 | 23.83 | 22.01 | 1.82 |
| 6131 | 6115 | 2.66 | 86.00 | 79.43 | 6.57 |
| 6302 | 6312 | 0.16 | 31.98 | 31.55 | 0.43 |
| 6312 | 6302 | 3.76 | 100.00 | 98.67 | 1.33 |
| 7292 | 7293 | 0.07 | 9.03 | 6.94 | 2.09 |
| 7293 | 7292 | 0.11 | 8.11 | 6.23 | 1.88 |
and if we look at a plot of one of these regions and how the overlap (y axis is % so between 0 and ~0.8%) changes with time
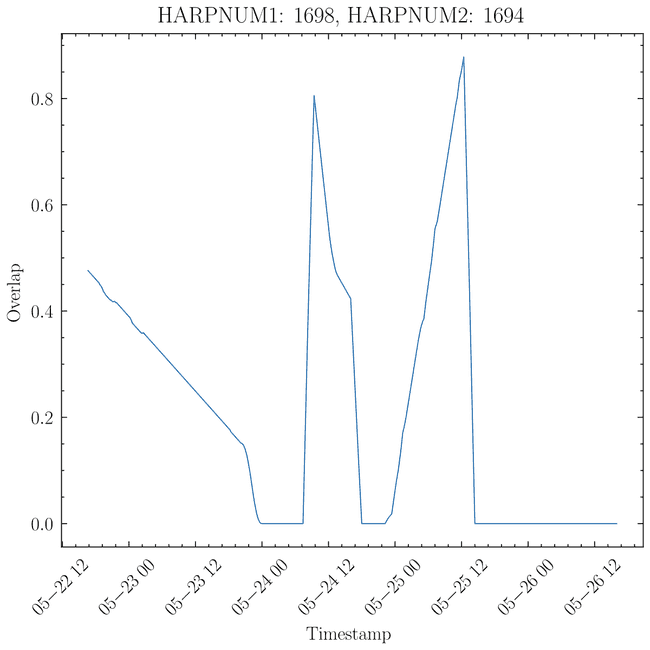
we see that indeed there are times when the overlap stops and then continues. From the table above, this seems to happen (intuitively) only for overlaps that are small to begin with. So, in practice, this isn't a big deal, but since for all other cases the result is the same (as there are no gaps in timestamps), and this is the real correct way to do it, we change to using
CREATE TEMP TABLE avg_overlap AS
SELECT tpo.harpnum1 as harpnum_a, tpo.harpnum2 as harpnum_b, AVG(tpo.overlap_percent) AS mean_overlap,
(100.0 * (COUNT(tpo.timestamp) - 1) * 12.0 * 60.0) / (1.0 * NULLIF(strftime('%s', H.end) - strftime('%s', H.start), 0)) AS ocurrence_percentage
FROM temp_overlap tpo
JOIN HARPS H ON H.harpnum = tpo.harpnum1
WHERE tpo.overlap_percent > 0
GROUP BY harpnum1, harpnum2;
CREATE TABLE OVERLAPS AS
SELECT ao.*,
-- Standard deviation of the overlap
SQRT(SUM((tpo.overlap_percent - ao.mean_overlap) * (tpo.overlap_percent - ao.mean_overlap)) / (COUNT(tpo.timestamp) - 1)) AS std_overlap
FROM avg_overlap ao
INNER JOIN TEMP_OVERLAP tpo ON ao.harpnum_a = tpo.harpnum1 AND ao.harpnum_b = tpo.harpnum2
GROUP BY harpnum_a, harpnum_b;substracting one because we don't count the last timestamp. Notice we also calculate the standard deviation of the overlap.
Aside: small overlaps
There are quite a considerable number of very small SHARP regions () of the solar surface. I'm not really sure what to do with them, so for now they stay. If anything, when it comes to training a ML model we can do undersampling of the mayority class (since I would not expect any of these regions to produce a CME) by removing this ones.
Managing overlaps
Once the overlaps are calculated, decisions need to be made as to what regions will be removed or merged (only a syntactic difference, as both involve removing one region). The rules for this are as follows
-
In order for an overlap to be considered problematic either
a. There's a >50% overlap with an occurence >50%.
b. There's a 100% overlap.
-
If any of those is fulfilled
-
If the occurence percentage is >70% and the average overlap is >90% then the region with the smaller area is removed and said to be "merged" with the larger one (as it's sufficiently similar to it).
-
Otherwise, the smaller region is said to be removed (as the overlap is significant but not enough to consider it a merged region).
-
In total 450 regions are removed. 147 are deleted and 303 are merged. With 430 having overlaps larger than 90%.
With the pre-processing steps finished, we can create the PROCESSED_HARPS_BBOX table which contains the bounding boxes that will be used for the rest of the analysis. This is done as follows
DROP TABLE IF EXISTS PROCESSED_HARPS_BBOX;
CREATE TABLE PROCESSED_HARPS_BBOX AS
SELECT RHB.* FROM RAW_HARPS_BBOX RHB
INNER JOIN HARPS H
ON RHB.harpnum = H.harpnum
WHERE H.area < 18 AND RHB.harpnum NOT IN (SELECT harpnum_a FROM OVERLAP_RECORDS);
DELETE FROM PROCESSED_HARPS_BBOX
WHERE (LONDTMIN < -90 AND LONDTMAX < -90)
OR (LONDTMIN > 90 AND LONDTMAX > 90);
UPDATE PROCESSED_HARPS_BBOX
SET LONDTMIN = -90
WHERE LONDTMIN < -90;
UPDATE PROCESSED_HARPS_BBOX
SET LONDTMAX = 90
WHERE LONDTMAX > 90;Associating CMEs to SHARP regions
Once the bounding boxes are processed, they can be used to associate CMEs to SHARP regions. This begins by running a series of scripts that perform the following (perhaps detailed code journey entry to come in the future)
-
Extracting the CMEs from the LASCO CME catalogue that have a quality of 0 (i.e. n poor quality comment).
-
Finding which SHARP regions were present at the time of each CME (temporally consistent) by using the start and end times of the SHARPs in the
PROCESSED_HARPS_BBOXtable. -
From the temporally consistent SHARP regions, for each CME, find which ones are spatially consistent. That is, the position angle of the SHARP region at the time of the CME should be withing the half the width of the CME plus five degrees (as an extra padding). Regions that are both spatially and temporally consistent are spatiotemporally consistent with a particular CME.
-
Match dimmings from solar demon (private communication). This is done by getting all the SHARP regions present at the time of the dimming's max detection time. Then, distances are calculated between the dimming's centroid and the SHARP regions' bounding boxes, with any point inside the bounding box being taken as distance 0. The SHARP with the closest distance is then associated to the dimming. If the closest distance is larger than 10 degrees, then the dimming is not associated to any SHARP region.
The reason we need to allow for some distance is because the dimming's centroid is calculated using the average of the pixels in the dimming such that if the dimming could be balanced on the position point, so an extended object is represented by a single point.
-
Extract flares from SWAN-SF. This is simply done by going through all the SWAN-SF files and recording any flare.
HOWEVER, due to overlaps requiring some SHARPs being removed, their flares are, in principle, lost. There are two ways to from here. Either accept that they're lost or transfer them to the region that prevailed. If doing the later, then it would be advisable to do this only for the regions that are "merged". Even then, it's true that we only store average values of the overlap, so it's possible that at the particular time of the flare, there's no overlap between the regions and we wrongly re-assign it.
Between the choice of losing some associations and wrongly assigning some, I think the former is better.
Loading the results into the database
We load the results into the database as follows
Spatially consistent
# Spatially consistent
df = pd.read_pickle(SPATIOTEMPORAL_MATCHING_HARPS_DATABASE_PICKLE)
df = df[df["HARPS_SPAT_CONSIST"]]
for cme_id, harpnum in df[["CME_ID", "HARPNUM"]].values:
cme_id = int(cme_id[2:])
harpnum = int(harpnum)
# Add the match to CMES_HARPS_SPATIALLY_CONSIST
new_cur.execute("""
INSERT INTO CMES_HARPS_SPATIALLY_CONSIST (harpnum, cme_id)
VALUES (?, ?)
""", (harpnum, cme_id))
# Dimmings
dimmings = pd.read_pickle(DIMMINGS_MATCHED_TO_HARPS_PICKLE)
dimmings = dimmings[dimmings["MATCH"]]
for i, row in dimmings[dimmings["MATCH"]].iterrows():
new_cur.execute(
"""
INSERT INTO dimmings (dimming_id, harpnum, harps_dimming_dist, dimming_start_date, dimming_peak_date, dimming_lon, dimming_lat)
VALUES (?, ?, ?, ?, ?, ?, ?)
""",
(
int(row["dimming_id"]),
int(row["HARPNUM"]),
row["HARPS_DIMMING_DISTANCE"],
row["start_time"].split(".")[0],
row["max_detection_time"].iso[:-4],
row["longitude"],
row["latitude"]
)
)
# Flares
flares = pd.read_pickle(FLARES_MATCHED_TO_HARPS_PICKLE)
for i, row in flares.iterrows():
new_cur.execute(
"""
INSERT INTO flares (flare_id, HARPNUM, flare_date, flare_lon, flare_lat, flare_class_score, flare_class, flare_ar, flare_ar_source, flare_verification)
VALUES (?, ?, ?, ?, ?, ?, ?, ?, ?, ?)
""",
(
int(row["FLARE_ID"]),
int(row["HARPNUM"]),
row["FLARE_DATE"].iso[:-4],
row["FLARE_LON"],
row["FLARE_LAT"],
row["FLARE_CLASS_SCORE"],
row["FLARE_CLASS"],
row["FLARE_AR"],
row["FLARE_AR_SOURCE"],
row["FLARE_VERIFICATION"]
)
)
Additionally, for each of these events (flares, dimmings and also cmes) we add an image_timestamp column which is closest timestamp with 00, 12, 24, 36 or 48 minutes. This is done as follows
def formatted_timestamp(timestamp):
"""
Format it to be the same hour and the closest minutes out
of 00, 12, 24, 36, 48
Using datetime
"""
minutes = [0, 12, 24, 36, 48]
# Check if already is a datetime
if not isinstance(timestamp, datetime):
datetime_timestamp = datetime.strptime(timestamp, "%Y-%m-%d %H:%M:%S")
else:
datetime_timestamp = timestamp
# Get the closest minute
minute = datetime_timestamp.minute
closest_minute = min(minutes, key=lambda x: abs(x - minute))
# Now we need to check if we need to add an hour
# We may need to if the original minute was greater than 48
if closest_minute == 0 and minute > 48:
# Add an hour
datetime_timestamp += timedelta(hours=1)
# Now we need to set the minute to the closest minute
datetime_timestamp = datetime_timestamp.replace(minute=closest_minute)
return datetime_timestamp
image_timestamps = new_cur.execute("SELECT DISTINCT(timestamp) FROM PROCESSED_HARPS_BBOX").fetchall()
image_timestamps = [t[0] for t in image_timestamps]
timestamps = sorted([datetime.strptime(t, "%Y-%m-%d %H:%M:%S") for t in image_timestamps])
image_timestamps = timestamps
# Dimmings
for row in tqdm(new_cur.execute("SELECT dimming_id, dimming_start_date FROM dimmings").fetchall()):
dimming_timestamp = datetime.strptime(row[1], "%Y-%m-%d %H:%M:%S")
image_timestamp = formatted_timestamp(dimming_timestamp)
new_cur.execute("UPDATE dimmings SET image_timestamp = ? WHERE dimming_id = ?", (image_timestamp.strftime("%Y-%m-%d %H:%M:%S"), row[0]))
# Same for flares
for row in tqdm(new_cur.execute("SELECT flare_id, flare_date FROM flares").fetchall()):
flare_timestamp = datetime.strptime(row[1], "%Y-%m-%d %H:%M:%S")
image_timestamp = formatted_timestamp(flare_timestamp)
new_cur.execute("UPDATE flares SET image_timestamp = ? WHERE flare_id = ?", (image_timestamp.strftime("%Y-%m-%d %H:%M:%S"), row[0]))
# And for cmes
for row in tqdm(new_cur.execute("SELECT cme_id, cme_date FROM cmes").fetchall()):
cme_timestamp = datetime.strptime(row[1], "%Y-%m-%d %H:%M:%S")
image_timestamp = formatted_timestamp(cme_timestamp)
new_cur.execute("UPDATE cmes SET image_timestamp = ? WHERE cme_id = ?", (image_timestamp.strftime("%Y-%m-%d %H:%M:%S"), row[0]))
new_conn.commit()Assigning events to a CME
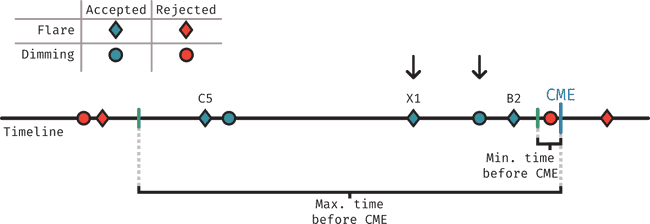
The figure above describes visually how events (flares and dimmings) are assigned to each CME. In short, the process works by considering a time period starting some time before the CME was detected in LASCO (min. tie before CME), to allow for the time that the CME takes to reach LASCO's FOV (using 12 minutes here to not be restrictive) and ending at some maximum time before the CME (here using 2 hours).
-
For dimmings: We find all events that fall within the time period defined above, then choose the closest one to the CME.
-
For flares: We find all the events that fall within the time period defined above, then choose the highest class flare.
Whenever one of the events is assigned to a CME, it's tracked as matched so it isn't considered in further CMEs.
Final associations and verification levels
Once events have been associated with their potential CMEs, we perform the final association between CMEs and SHARP regions. To do so, we define five levels of verification (meant to indicate the level of confidence, specially for level 1, but we remark that this is an subjective classification). All of these levels refer to spatiotemporally consistent SHARP regions. These are
-
Level 1: A region where a dimming and a flare of class larger than C5 can be matched to the CME.
-
Level 2: A region where a flare with class larger than C5 can be matched to the CME but not a dimming.
-
Level 3: A region where a dimming and a flare of class smaller than C5 can be matched to the CME.
-
Level 4: A region where a flare with class smaller than C5 can be matched to the CME but not a dimming.
-
Level 5 A region where a dimming can be matched to the CME but not a flare.
Or in table format
| Level | Flare | Dimming |
|---|---|---|
| 1 | C5+ | Yes |
| 2 | C5+ | No |
| 3 | C5- | Yes |
| 4 | C5- | No |
| 5 | No | Yes |
In order to perform the final matching, we select all the candidate SHARP regions for each CME and then select the one with the highest verification level. This is done as follows
unique_cmes = new_cur.execute("SELECT DISTINCT cme_id from CMES_HARPS_SPATIALLY_CONSIST").fetchall()
# Now let's get the matches
def get_verfification_level(has_dimming, has_flare, flare_class, flare_threshold=25):
if has_dimming:
if has_flare:
if flare_class > flare_threshold:
return 1
else:
return 3
else:
return 5
else:
if has_flare:
if flare_class > flare_threshold:
return 2
else:
return 4
else:
return -1
def verif_level_from_row(row):
has_dimming = row['has_dimming']
has_flare = 0 if pd.isnull(row['flare_id']) else 1
flare_class = None if pd.isnull(row['flare_class_score']) else row['flare_class_score']
harpnum = row['harpnum']
return get_verfification_level(has_dimming, has_flare, flare_class)
matches = dict()
for unique_cme in tqdm(unique_cmes):
query = f"""
SELECT CHSC.cme_id, CHSC.harpnum, CHE.flare_id, CHE.dimming_id, F.flare_class_score from CMES_HARPS_SPATIALLY_CONSIST CHSC
LEFT JOIN CMES_HARPS_EVENTS CHE ON CHSC.cme_id = CHE.cme_id AND CHSC.harpnum = CHE.harpnum
LEFT JOIN FLARES F ON CHE.flare_id = F.flare_id
WHERE CHSC.cme_id = {unique_cme[0]}
"""
df = pd.read_sql_query(query, new_conn)
# Need to replace dimming_id here with either 0 or 1
df['has_dimming'] = df['dimming_id'].apply(lambda x: 0 if pd.isnull(x) else 1)
sorted_df = df.sort_values(by=['has_dimming', 'flare_class_score'], ascending=False)
# Apply verif_level_from_row to each row
df["verif_level"] = df.apply(verif_level_from_row, axis=1)
sorted_df = df.sort_values(by=['verif_level'], ascending=True)
# But we don't want verification level -1
sorted_df = sorted_df[sorted_df['verif_level'] != -1]
# And if this is empty, well there's no match so we continue
if sorted_df.empty:
continue
# Otherwise we can continue
top_choice = sorted_df.iloc[0]
verification_level = top_choice['verif_level']
harpnum = top_choice['harpnum']
# There's a match!
matches[unique_cme[0]] = {
'harpnum': harpnum,
'verification_level': verification_level
}
new_cur.execute("DELETE FROM FINAL_CME_HARP_ASSOCIATIONS")
# Iterate through key, value pairs of matches
for cme_id, values in tqdm(matches.items()):
harpnum = int(values["harpnum"])
verification_score = int(values["verification_level"])
association_method = "automatic"
independent_verfied = 0
# Add to database
new_cur.execute("INSERT INTO FINAL_CME_HARP_ASSOCIATIONS (cme_id, harpnum, verification_score, association_method, independent_verified) VALUES (?, ?, ?, ?, ?)", (cme_id, harpnum, verification_score, association_method, independent_verfied))
new_conn.commit()Data reduction summary
The following table summarises how the number of CMEs and SHARP regions has changed through the different steps taken here
| Step | Substep | CMEs | SHARP regions |
|---|---|---|---|
| Starting point | 32684 | 4098 | |
| Pre-processing | Removing big harps | 32684 | 4096 |
| Pre-processing | Removing overlaps | 32684 | 3655 |
| Matching | CMEs during HARPs exist | 14507 | 3655 |
| Matching | Spatiotemporally matching regions | 3755 | 3655 |
| Matching | Match criteria | 1001 | 3655 |
