Dataset II - Building a general CME sequences dataset
The purpose of this code journey is to build, based on the work of building a CME source region catalogue, a general dataset consisting of sliding sequences in time of SHARP regions, labelled according to how long after the end of an observation period a CME (if any) occurs.Defining the sequences
Sequences were originally designed to be formed of a lead-in , observation and forecasting periods
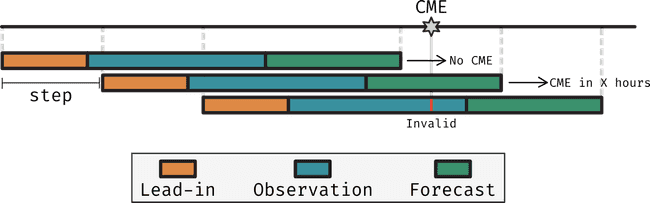
- Lead-in period: Period before the observation period, where events occurring in this period could influence what's seen in the observation period
- Observation period: The period where data, whatever form it takes, is used to make predictions.
- Forecasting period: The period after the observation period, where the prediction is made.
However, in discussions with the team at Dublin DIAS, it was mentioned that neural networks may have a hard time when asked to forecast a binary label (yes/no CME) in such a form because
- A CME happening just before the observation period may not be seen in the observation period, but may still be relevant to the prediction. While we wanted to capture this in the lead-in period there's nothing we do about it.
- A CME just after the end of the observation period may produce signatures in the observation period that are picked up by the neural network, resulting in conflicting information for the neural network to learn from. It is seeing signatures of a CME yet we tell it the label is no CME.
So, instead of defining sequences according to these three periods, we will define them according only to the observation period. We will then label the sequence by how long after the observation period did a CME (if any) will occur. Additionally, we will record the number of CMEs that happened before a particular sequence and how long ago the last one happened. This extra information about the past of the region may then be used as extra input to the model.

Extracting sequences from the data
The sequences are then defined by two quantities:
- Length of the observation period (O): The length of the observation period in hours.
- Step (S): The step in hours between the start of each observation period.
- The start of the first observation period: Which can either be the start of the region's lifetime or e.g. when it falls completely within (-70,70) degrees longitude.
- The last observation period: Which can either be the end of the region's lifetime or e.g. when it falls completely within (-70,70) degrees longitude.
Now, other aspects to consider are e.g. which CMEs will be taken into consideration. We must not forget that it's likely that CMEs associated with regions when they were closer to the limb of the Sun are more likely to be misassociations with a real region producing the CME in the back of the Sun. The labels of the sequences depend on what we decide to do with this.
Sequences boundaries
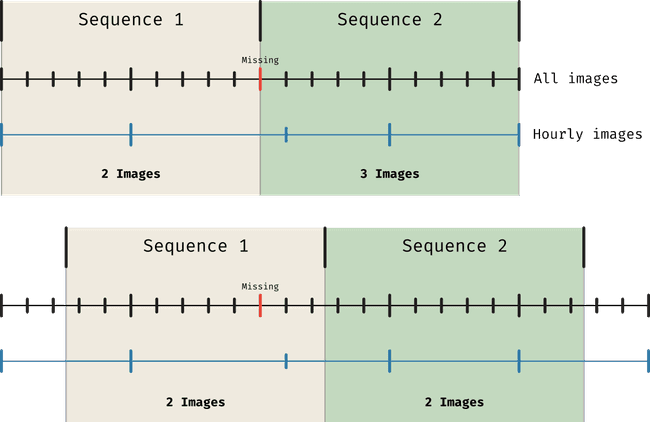
Although here we're focusing on a general dataset (not really a dataset but still) that is not constrained by the limitations of some data source, we could define the boundaries of the sequences to be any time. However, keeping in mind that later on we will be using SDOML data, and that it's not a bad idea, whatever the data is, to keep a cadence of one hour, we must be aware of the limitations this will bring. In particular, if our data source is missing some entries (like SDOML does), we may want to mitigate that by choosing (say we take the first period to start a o'clock) entries that differ a bit from the timestamp we wanted (we may take 12 or 24 minutes past the hour). This will lead to problems if we choose for both our slices to be at o'clock and we downsample our data to be one entry at every o'clock.
In simple terms, when we allow any of :00, :12 or :24 to be considered as our images, we want to make sure that our sequence splits don't start anywhere that will break the :00 to :24 period. If that happens at the end of a sequence, we may find that if the last entry of the sequence is missing say :00 and has been taken up by :12, if our sequence ends at :00 we will be missing this last image even though it's available. So, instead of having sequences starting at :00 we can have them starting at :30, resolving this problem. In the figure below we illustrate this issue. The top part of the figure shows what happens when the timestamps we choose for the hourly images coincides with the edges of the sequences. The number of images in each sequence is not consistent. In the bottom part we show how this is solved by simply adding a 30min offset to the start of the sequences.
How the dataset is built
The basic idea for the dataset is to have a python class that represents a single harpnum and at each step it processes one slice before moving by the step. Then, what we want is to get the first and last timestamps for each sequence and check the observation, lead-in and prediction periods for each slice. As an output for each step we should get a row that is either accepted or rejected (if say, there's a final association CME in the observation period).
Which bounding boxes to use?
As we mentioned before, we need to choose whether we want to use the full lifetime of HARPs regions, or whether we want to use the time when they are within (-70,70) degrees longitude.
Now, magnetic field information is essential for processes in the Sun, specially for CMEs, and it's common in the literature to consider that magnetic field information (including magnetograms) too close to the limb may not be reliable. Given this, we choose to only consider the times when a SHARP region was within (-70,70) degrees longitude for the dataset. So we create a table NO_LIMB_BBOX just like this. In the function we also create some indices that speed up the process.
def create_temp_table() -> None:
conn = sqlite3.connect(CMESRCV3_DB)
cur = conn.cursor()
cur.execute("DROP TABLE IF EXISTS NO_LIMB_BBOX")
cur.execute("""
CREATE TABLE NO_LIMB_BBOX AS
SELECT * FROM PROCESSED_HARPS_BBOX
WHERE LONDTMIN > -70 AND LONDTMAX < 70
""")
cur.executescript("""
CREATE INDEX IF NOT EXISTS idx_no_limb_bbox_harpnum ON NO_LIMB_BBOX(harpnum);
CREATE INDEX IF NOT EXISTS idx_no_limb_bbox_timestamp ON NO_LIMB_BBOX(timestamp);
CREATE INDEX IF NOT EXISTS idx_no_limb_bbox_harpnum_timestamp ON NO_LIMB_BBOX(harpnum, timestamp);
CREATE INDEX IF NOT EXISTS idx_no_limb_bbox_londtmin ON NO_LIMB_BBOX(latdtmin);
CREATE INDEX IF NOT EXISTS idx_no_limb_bbox_londtmax ON NO_LIMB_BBOX(latdtmax);
CREATE INDEX IF NOT EXISTS idx_fcha_harpnum ON FINAL_CME_HARP_ASSOCIATIONS(harpnum);
CREATE INDEX IF NOT EXISTS idx_cmes_date ON CMES(cme_date);
""")
conn.commit()
conn.close()The class
We define the skeleton of the class as follows
class HarpsDatasetSlices():
def __init__(self, harpnum: int, O: Quantity, S: Quantity, db_path: str, strict: bool, table: str):
# Initialise the class
pass
def __check_period_lengths(self) -> None:
# Checking the input period lengths are correct
pass
def __get_first_timestamp(self) -> astropy.time.Time:
# Getting the first timestamp of the HARP's lifetime
pass
def __get_last_timestamp(self) -> astropy.time.Time:
# Getting the last timestamp of the HARP's lifetime
pass
def _get_period_bounds(self) -> None:
# Getting the bounds of the observation period
# and the lead-in and prediction periods
pass
def _check_observation_period(self) -> int:
# Checking that during the observation period
# there's no final association CME
# or if strict is True, also that the region
# wasn't spatiotemporally consistent with a CME
# with which it wasn't associated but it had
# a flare or dimming that could be associated
# with it (to remove ambiguous cases)
pass
def _get_previous_cme(self) -> Union[Tuple[None,None, int], Tuple[int, float, int]]:
# Getting the last CME that happened before the
# observation period as well as the number of
# all CMEs that happened before the observation period
pass
def _get_label(self) -> Tuple[int, Union[float, None], Union[int, None], Union[int, None]]:
# Getting the label for the sequence,
# either 0 if no CMEs in the future or
# 1 + the number of hours until the next CME
# if there's a CME in the future
pass
def get_current_row(self) -> Tuple[int, Union[accepted_row, rejected_row]]:
# Processing the current slice and returning
# a row with the information
pass
def step(self) -> None:
# Moving the class to the next slice
pass
We now discuss each of the methods in detail.
__init__
def __init__(self, harpnum: int, O: Quantity, S: Quantity, db_path: str = CMESRCV3_DB, strict: bool = False, table: str = "PROCESSED_HARPS_BBOX"):
"""
Initialize a HarpsDatasetSlices object to manage HARPS dataset slices for solar physics research.
Parameters
----------
harpnum : int
The HARP number of the dataset to be analyzed.
O : Quantity
The length of the observation period. Must be an Astropy Quantity object with time units.
S : Quantity
The step size for moving the observation window. Must be an Astropy Quantity object with time units.
db_path : str, optional
The path to the database containing HARPS data. Default is `CMESRCV3_DB`.
strict : bool, optional
Whether to enforce strict conditions for the observation period. Default is False.
table : str, optional
The name of the database table to query for data. Default is "PROCESSED_HARPS_BBOX".
Attributes
----------
table : str
The name of the database table to use.
harpnum : int
The HARP number of the dataset.
O : float
The length of the observation period in hours.
S : float
The step size in hours.
strict : bool
Whether to enforce strict conditions for the observation period.
conn : sqlite3.Connection
The SQLite database connection.
cur : sqlite3.Cursor
The SQLite database cursor for executing SQL queries.
first_timestamp : datetime
The first timestamp of the dataset.
last_timestamp : datetime
The last timestamp of the dataset.
current_timestamp : datetime
The current timestamp for slicing.
lead_in_period : tuple
The start and end times of the lead-in period.
observation_period : tuple
The start and end times of the observation period.
prediction_period : tuple
The start and end times of the prediction period.
finished : bool
Whether the dataset has been fully processed.
"""
# Initialize basic attributes
self.table = table
self.harpnum = int(harpnum)
self.O = O.to(u.hour).value
self.S = S.to(u.hour).value
self.strict = strict
# Check validity of period lengths
self.__check_period_lengths()
# Initialize database connection and cursor
self._initialize_db(db_path)
# Fetch the first and last timestamps
self.first_timestamp, self.last_timestamp = self.__get_timestamp_bounds()
# Normalize the timestamps
self._normalize_timestamps()
# Initialize other attributes
self.current_timestamp = self.first_timestamp
self.lead_in_period = None
self.observation_period = None
self.prediction_period = None
self.finished = False
# Compute initial period bounds
self._get_period_bounds()__init__ simply initializes the class. We assign the table from which data is obtained, the value of the harpnum, the length of the observation period and the step and whether we will be doing strict checks or not. We then check the validity of the period lengths using __check_period_lengths()
def __check_period_lengths(self) -> None:
"""
Validate the lengths of the observation and step periods.
This method checks if the lengths of the observation period (`O`) and the step size (`S`)
are both positive and multiples of 1 hour. Raises a `ValueError` if any of these conditions
are not met.
Raises
------
ValueError
If the observation period or step size is either negative or not a multiple of 1 hour.
Returns
-------
None
"""
# Define a mapping between attribute names and their human-readable equivalents
period_names = {
'O': "Observation period",
'S': "Step size"
}
# Validate each period length
for period_value, period_name in zip([self.O, self.S], period_names.keys()):
if period_value < 0:
raise ValueError(f"{period_names[period_name]} must be positive.")
if period_value % 1 != 0:
raise ValueError(f"{period_names[period_name]} must be a multiple of 1 hour.")we also initialize a connection to the database here using _initialize_db
def _initialize_db(self, db_path: str):
"""
Initialize the database connection and cursor.
"""
self.conn = sqlite3.connect(db_path)
self.conn.execute("PRAGMA foreign_keys = ON")
self.cur = self.conn.cursor()get the first and last timestamps for the HARPs region
def __get_timestamp_bounds(self) -> Tuple[astropy.time.Time, astropy.time.Time]:
"""
Get the first and last timestamps for the given harpnum.
This method fetches the earliest and latest timestamps for the harpnum from the database.
It raises a specific error if no data is found for the given harpnum.
Raises
------
NoBBoxData
If no bounding box data is available for the given harpnum.
Returns
-------
Tuple[astropy.time.Time, astropy.time.Time]
A tuple containing the first and last timestamps as astropy.time.Time objects.
"""
# Fetch the first timestamp
self.cur.execute(
f"SELECT MIN(datetime(timestamp)) FROM {self.table} WHERE harpnum = ?", (self.harpnum,)
)
first_str_timestamp = self.cur.fetchone()[0]
# Fetch the last timestamp
self.cur.execute(
f"SELECT MAX(datetime(timestamp)) FROM {self.table} WHERE harpnum = ?", (self.harpnum,)
)
last_str_timestamp = self.cur.fetchone()[0]
# Check if either timestamp is None
if first_str_timestamp is None or last_str_timestamp is None:
raise NoBBoxData()
# Convert to datetime objects
first_timestamp = datetime.fromisoformat(first_str_timestamp)
last_timestamp = datetime.fromisoformat(last_str_timestamp)
return first_timestamp, last_timestampand following the discussion above, normalize these timestamps to be at half past hours using self._normalize_timestamps()
def _normalize_timestamps(self):
"""
Normalize the first and last timestamps to be at half past the hour.
"""
self.first_timestamp = self.first_timestamp.replace(minute=30, second=0)
self.last_timestamp = self.last_timestamp.replace(minute=30, second=0)Finally, we initialize some other attributes
# Initialize other attributes
self.current_timestamp = self.first_timestamp
self.lead_in_period = None
self.observation_period = None
self.prediction_period = None
self.finished = Falseand end by getting the bounds of the different periods using _get_period_bounds
def _get_period_bounds(self) -> None:
"""
Compute and set the bounds for the lead-in, observation, and prediction periods.
This method calculates the bounds for the lead-in, observation, and prediction periods based
on the current timestamp, observation period length (`O`), first timestamp, and last timestamp.
Sets the attributes `lead_in_period`, `observation_period`, and `prediction_period` with the
calculated bounds.
Returns
-------
None
"""
# Pre-compute end of observation period based on current timestamp and observation length
end_of_observation = self.current_timestamp + timedelta(hours=self.O)
# Calculate bounds for each period
lead_in_bounds = (self.first_timestamp, self.current_timestamp)
observation_bounds = (self.current_timestamp, end_of_observation)
prediction_bounds = (end_of_observation, self.last_timestamp)
# Assign calculated bounds to class attributes
self.lead_in_period = lead_in_bounds
self.observation_period = observation_bounds
self.prediction_period = prediction_bounds_check_observation_period
def _check_observation_period(self) -> None:
"""
Validate the current observation period based on certain conditions.
This method checks if the current observation period is valid by querying the database.
It raises an `InvalidObservationPeriod` exception if the observation period is invalid
based on one of two conditions:
1. If `strict` is True, it checks whether a CME event is associated with a dimming or flare.
2. Checks for a final CME association.
Raises
------
InvalidObservationPeriod
If the observation period is invalid based on the criteria.
Returns
-------
None
"""
# Extract and format start and end dates of the observation period
start_dt, end_dt = self.observation_period
start = (start_dt - timedelta(seconds=1)).strftime("%Y-%m-%d %H:%M:%S")
end = (end_dt + timedelta(seconds=1)).strftime("%Y-%m-%d %H:%M:%S")
# Check for CME events if 'strict' is True
if self.strict:
self._check_strict_condition(start, end)
# Check for final CME association
self._check_final_cme_association(start, end)
def _check_strict_condition(self, start: str, end: str) -> None:
"""Check if region was spatiotemporally consistent with a CME and if so, whether any dimming or flares are associated."""
query = """
SELECT CHE.cme_id, CHE.flare_id, CHE.dimming_id FROM CMES_HARPS_EVENTS CHE
JOIN CMES C ON CHE.cme_id = C.cme_id
JOIN CMES_HARPS_SPATIALLY_CONSIST CHSC ON CHSC.harpnum = CHE.harpnum AND CHSC.cme_id = CHE.cme_id
WHERE ((CHE.dimming_id NOT NULL) OR (CHE.flare_id NOT NULL))
AND CHE.harpnum = ?
AND datetime(C.cme_date) BETWEEN datetime(?) AND datetime(?)
AND CHE.cme_id NOT IN (SELECT cme_id FROM FINAL_CME_HARP_ASSOCIATIONS WHERE harpnum = ?)
"""
self.cur.execute(query, (self.harpnum, start, end, self.harpnum))
results = self.cur.fetchall()
if len(results) != 0:
raise InvalidObservationPeriod("Observation period has a CME with dimming or flare associated with it.",
reason='unclear_cme_present',
dimming_id=results[0][2],
flare_id=results[0][1],
cme_id=results[0][0])
def _check_final_cme_association(self, start: str, end: str) -> None:
"""Check for a final CME association."""
query = """
SELECT FCHA.cme_id FROM FINAL_CME_HARP_ASSOCIATIONS FCHA
JOIN CMES C ON C.cme_id = FCHA.cme_id
WHERE FCHA.harpnum = ?
AND datetime(C.cme_date) BETWEEN datetime(?) AND datetime(?)
"""
self.cur.execute(query, (self.harpnum, start, end))
results = self.cur.fetchall()
if len(results) != 0:
raise InvalidObservationPeriod("Observation period has a final CME association.",
reason='final_cme_association',
cme_id=results[0][0])Here the aim is to check whether
- If we're in strict mode, the region was spatiotemporally consistent with a CME and if so, where there any dimming or flares that could've been associated with it?
- If there's a final CME association in the observation period.
If any of those is true, the sequence isn't valid and we raise an InvalidObservationPeriod exception. These exceptions will be handled later on when creating rows to distinguish between accepted and rejected rows.
_get_previous_cme
def _get_previous_cme(self) -> Union[Tuple[None, None, int], Tuple[int, float, int]]:
"""
Get the previous CME information for the given harpnum.
This method queries the database to find CME events that occurred during the lead-in period
for the given harpnum. It returns information about the closest CME to the start of the observation
period, as well as the total number of CME events during the lead-in period.
Raises
------
ValueError
If no lead-in period is available.
Returns
-------
Union[Tuple[None, None, int], Tuple[int, float, int]]
A tuple containing:
- The closest CME ID (or None if no CMEs).
- The time difference between the start of the observation period and the closest CME (or None).
- The total number of CMEs in the lead-in period.
"""
# Validate lead-in period
if self.lead_in_period is None:
raise ValueError("No lead-in period.")
# Prepare time bounds for query
start = self.lead_in_period[0].strftime("%Y-%m-%d %H:%M:%S")
end = self.lead_in_period[1].strftime("%Y-%m-%d %H:%M:%S")
# Query database for CMEs during the lead-in period
query = """
SELECT FCHA.cme_id, C.cme_date, FCHA.verification_score FROM FINAL_CME_HARP_ASSOCIATIONS FCHA
JOIN CMES C ON C.cme_id = FCHA.cme_id
WHERE FCHA.harpnum = ?
AND datetime(C.cme_date) BETWEEN datetime(?) AND datetime(?)
ORDER BY datetime(C.cme_date) DESC
"""
self.cur.execute(query, (self.harpnum, start, end))
results = self.cur.fetchall()
counts = {level: 0 for level in range(1, 6)}
# Check if any CMEs were found
if len(results) == 0:
return (None, None, 0, counts)
for _, _, level in results:
counts[level] += 1
# Extract closest CME and calculate time difference
cme_id = int(results[0][0])
cme_date = datetime.fromisoformat(results[0][1])
diff = float((self.observation_period[0] - cme_date).total_seconds() / 3600)
return (cme_id, diff, len(results), counts)The purpose of this function is to count how many CMEs happened before the start of the observation period (both total number but also by verification level) as well as returning the time difference between the start of the observation period and the closest CME (and its cme_id). This information could become relevant in the training of the model as outlined at the beginning of this post.
_get_label
def _get_label(self) -> Tuple[int, Union[float, None], Union[int, None], Union[int, None]]:
"""
Get the label and relevant metadata for the current observation period.
This method queries the database to find CME events that occurred during the prediction period
for the given harpnum. It returns information about the CME closest to the end of the observation
period, as well as its verification level.
Returns
-------
Tuple[int, Union[float, None], Union[int, None], Union[int, None]]
A tuple containing:
- Binary label indicating if a CME occurred (1) or not (0).
- Time until the closest CME in hours (or None if no CMEs).
- Verification level of the closest CME (or None).
- ID of the closest CME (or None).
"""
# Prepare time bounds for query
start = self.prediction_period[0].strftime("%Y-%m-%d %H:%M:%S")
end = self.prediction_period[1].strftime("%Y-%m-%d %H:%M:%S")
# Query database for CMEs during the prediction period
query = """
SELECT FCHA.cme_id, C.cme_date, FCHA.verification_score FROM FINAL_CME_HARP_ASSOCIATIONS FCHA
JOIN CMES C ON C.cme_id = FCHA.cme_id
WHERE FCHA.harpnum = ?
AND datetime(C.cme_date) BETWEEN datetime(?) AND datetime(?)
ORDER BY datetime(C.cme_date) ASC
"""
self.cur.execute(query, (self.harpnum, start, end))
results = self.cur.fetchall()
# Check if any CMEs were found
if len(results) == 0:
return (0, None, None, None)
# Extract closest CME and calculate time difference
cme_id = int(results[0][0])
cme_date = datetime.fromisoformat(results[0][1])
diff = float((cme_date - self.observation_period[1]).total_seconds() / 3600)
verification_level = int(results[0][2])
return (1, diff, verification_level, cme_id)The purpose of this function is to check if there'll be any CMEs in the prediction period and if so return how long after the end of the observation period, what the verification level of that CME is and its cme_id. If no CMEs are found, we return 0 as the label.
get_current_row
def get_current_row(self) -> Tuple[int, Union[accepted_row, rejected_row]]:
"""
Retrieve the current row data for the dataset.
Returns
-------
Tuple[int, Union[accepted_row, rejected_row]]
A tuple containing a flag (1 for accepted row, 0 for rejected) and the row data.
"""
# Validate the observation period
try:
self._check_observation_period()
except InvalidObservationPeriod as e:
# Build and return the rejected row
return (
0, # Flag for a rejected row
(
self.harpnum,
self.lead_in_period[0].strftime('%Y-%m-%d %H:%M:%S'),
self.lead_in_period[1].strftime('%Y-%m-%d %H:%M:%S'),
self.observation_period[0].strftime('%Y-%m-%d %H:%M:%S'),
self.observation_period[1].strftime('%Y-%m-%d %H:%M:%S'),
self.prediction_period[0].strftime('%Y-%m-%d %H:%M:%S'),
self.prediction_period[1].strftime('%Y-%m-%d %H:%M:%S'),
e.reason,
e.message
)
)
# Get previous CME information and number of CMEs in the lead-in period
prev_cme_id, prev_diff, n_cmes, counts = self._get_previous_cme()
# Get the label for the observation period
label, diff, verification_level, cme_id = self._get_label()
# Build and return the accepted row
return (
1, # Flag for an accepted row
(
self.harpnum,
self.lead_in_period[0].strftime('%Y-%m-%d %H:%M:%S'),
self.lead_in_period[1].strftime('%Y-%m-%d %H:%M:%S'),
self.observation_period[0].strftime('%Y-%m-%d %H:%M:%S'),
self.observation_period[1].strftime('%Y-%m-%d %H:%M:%S'),
self.prediction_period[0].strftime('%Y-%m-%d %H:%M:%S'),
self.prediction_period[1].strftime('%Y-%m-%d %H:%M:%S'),
prev_cme_id,
prev_diff,
n_cmes,
counts[1],
counts[2],
counts[3],
counts[4],
counts[5],
label,
diff,
verification_level,
cme_id
)
)All of the previous methods are combined in _get_current_row to return a row with all the information we need. If the observation period is invalid, we return a rejected row with the reason why it was rejected. Otherwise, we return an accepted row with all the information we need.
step
def step(self) -> None:
"""
Step the current timestamp forward by S.
Returns
-------
None
"""
# Check we haven't reached the end of the dataset
if self.finished:
raise Finished("Dataset has been fully processed.")
next_observation_end = self.observation_period[1] + timedelta(hours=self.S)
if next_observation_end > self.last_timestamp:
self.finished = True
return
self.current_timestamp += timedelta(hours=self.S)
self._get_period_bounds()Finally, step simply moves the class forward by the step size and updates the period bounds. If after moving forward the observation period end is after the last timestamp, we set self.finished to True to indicate that we're done. With this implementation, self.finished=True doesn't stop anything in the class, but later on we use this flag to stop processing.
Other considerations
Additionally, outside the class we implement the following function
def get_harpnum_list() -> List[int]:
"""
Get the list of unique harp numbers from the database.
Returns
-------
List[int]
List of unique harp numbers.
"""
with sqlite3.connect(CMESRCV3_DB) as conn:
cur = conn.cursor()
cur.execute("SELECT DISTINCT harpnum FROM PROCESSED_HARPS_BBOX")
harpnum_list = [int(row[0]) for row in cur.fetchall()]
return harpnum_listto get all the harpnums that need to be processed. We also define the following exceptions that were used above
class Finished(Exception):
"""
Raised when the data processing is finished.
"""
pass
class InvalidObservationPeriod(Exception):
"""
Raised when an observation period is invalid.
Parameters
----------
message : str
The error message.
reason : str
The reason for invalidity. Must be one of ['missing_images', 'unclear_cme_present', 'final_cme_association'].
dimming_id : str, optional
The ID of the dimming event, required if reason is 'unclear_cme_present'.
flare_id : str, optional
The ID of the flare event, required if reason is 'unclear_cme_present'.
cme_id : str, optional
The ID of the CME event.
"""
def __init__(self, message: str, reason: str, dimming_id: str = None, flare_id: str = None, cme_id: str = None) -> None:
valid_reasons = ['missing_images', 'unclear_cme_present', 'final_cme_association']
if reason not in valid_reasons:
raise ValueError(f"Invalid reason {reason}.")
self.message = message
self.reason = reason
if self.reason == "unclear_cme_present":
if (dimming_id is None and flare_id is None) or cme_id is None:
raise ValueError("Must provide at least one of dimming_id, flare_id and a cme_id.")
self.dimming_id = dimming_id
self.flare_id = flare_id
self.cme_id = cme_id
if self.reason == "final_cme_association":
if cme_id is None:
raise ValueError("Must provide a cme_id.")
self.cme_id = cme_id
class NoBBoxData(Exception):
"""
Raised when no bounding box data is available.
"""
passProcessing all HARPs regions
In order to aid in the processing of all harpnums we also write the get_all_rows function (not part of the class) as follows
def get_all_rows(O: Quantity, S: Quantity, strict: bool = False, table: str = "PROCESSED_HARPS_BBOX") -> Tuple[List[AcceptedRow], List[RejectedRow]]:
"""
Generate all rows for a set of HARPs datasets.
Parameters
----------
O : Quantity
The observation period length. Astropy units required.
S : Quantity
The step size. Astropy units required.
strict : bool, optional
Whether to enforce strict criteria for observation periods. Default is False.
table : str, optional
Name of the table in the database. Default is "PROCESSED_HARPS_BBOX".
Returns
-------
Tuple[List[AcceptedRow], List[RejectedRow]]
A tuple containing two lists: one for accepted rows and one for rejected rows.
"""
# Get the list of HARPs numbers
harpnum_list = get_harpnum_list()
# Initialize lists for accepted and rejected rows
accepted_rows = []
rejected_rows = []
# Loop through each HARPs number
for harpnum in tqdm(harpnum_list):
try:
# Initialize the dataset
dataset = HarpsDatasetSlices(harpnum, O, S, strict=strict, table=table)
first_row = dataset.get_current_row()
except NoBBoxData:
# Handle cases with no bounding box data
first_row = (0, (harpnum, None, None, None, None, None, None, "no_bbox_data", "No BBox data for this harpnum."))
rejected_rows.append(first_row)
continue
# Add the first row to the appropriate list
accepted_rows.append(first_row) if first_row[0] == 1 else rejected_rows.append(first_row)
# Step through the dataset and collect rows
dataset.step()
while not dataset.finished:
row = dataset.get_current_row()
accepted_rows.append(row) if row[0] == 1 else rejected_rows.append(row)
dataset.step()
return accepted_rows, rejected_rowsThis function simply loops through all the harpnums, initializes the class for each one and collects the rows. It returns two lists, one for accepted rows and one for rejected rows.
Writing into a database
def write_into_database(accepted_rows: List[AcceptedRow], rejected_rows: List[RejectedRow], db_path: str = GENERAL_DATASET, main_database = CMESRCV3_DB) -> None:
"""
Write the accepted and rejected rows into the specified SQLite database.
Parameters
----------
accepted_rows : List[AcceptedRow]
List of accepted rows to be inserted into the GENERAL_DATASET table.
rejected_rows : List[RejectedRow]
List of rejected rows to be inserted into the GENERAL_DATASET_REJECTED table.
db_path : str, optional
The path to the SQLite database. Default is CMESRCV3_DB.
Returns
-------
None
"""
conn = sqlite3.connect(db_path)
conn.execute("PRAGMA foreign_keys = ON")
cur = conn.cursor()
if main_database != db_path:
conn.execute("ATTACH DATABASE ? AS CMESRCV3", (main_database,))
# Create the HARPS table, copying from the main database
cur.execute("DROP TABLE IF EXISTS main.HARPS")
cur.execute("""
CREATE TABLE main.HARPS AS
SELECT * FROM CMESRCV3.HARPS
""")
# Same with CMEs table
cur.execute("DROP TABLE IF EXISTS main.CMES")
cur.execute("""
CREATE TABLE main.CMES AS
SELECT * FROM CMESRCV3.CMES
""")
cur.execute("DROP TABLE IF EXISTS main.FINAL_CME_HARP_ASSOCIATIONS")
cur.execute("""
CREATE TABLE main.FINAL_CME_HARP_ASSOCIATIONS AS
SELECT * FROM CMESRCV3.FINAL_CME_HARP_ASSOCIATIONS
""")
# Detach the main database
conn.commit()
conn.execute("DETACH DATABASE CMESRCV3")
conn.commit()
# Create the accepted_rows table
cur.execute("DROP TABLE IF EXISTS GENERAL_DATASET")
cur.execute("""
CREATE TABLE IF NOT EXISTS GENERAL_DATASET (
slice_id INTEGER PRIMARY KEY AUTOINCREMENT,
harpnum INT NOT NULL,
lead_in_start TEXT NOT NULL,
lead_in_end TEXT NOT NULL,
obs_start TEXT NOT NULL,
obs_end TEXT NOT NULL,
pred_start TEXT NOT NULL,
pred_end TEXT NOT NULL,
prev_cme_id INT,
prev_cme_diff REAL,
n_cmes_before INTEGER NOT NULL,
n_cmes_before_1 INTEGER NOT NULL,
n_cmes_before_2 INTEGER NOT NULL,
n_cmes_before_3 INTEGER NOT NULL,
n_cmes_before_4 INTEGER NOT NULL,
n_cmes_before_5 INTEGER NOT NULL,
label INTEGER NOT NULL,
cme_diff REAL,
verification_level INTEGER,
cme_id INT
)
""")
# Create the rejected_rows table
cur.execute("DROP TABLE IF EXISTS GENERAL_DATASET_REJECTED")
cur.execute("""
CREATE TABLE IF NOT EXISTS GENERAL_DATASET_REJECTED (
slice_id INTEGER PRIMARY KEY AUTOINCREMENT,
harpnum INT NOT NULL,
lead_in_start TEXT,
lead_in_end TEXT,
obs_start TEXT,
obs_end TEXT,
pred_start TEXT,
pred_end TEXT,
reason TEXT NOT NULL,
message TEXT NOT NULL
)
""")
# Insert the rows into the tables
for row in tqdm(accepted_rows):
# Make double sure harpnum is int
# Also prev_cme_id and cme_id
nrow = list(row[1])
nrow[0] = int(nrow[0])
nrow = tuple(nrow)
try:
cur.execute("""
INSERT INTO GENERAL_DATASET
(harpnum, lead_in_start, lead_in_end, obs_start, obs_end, pred_start, pred_end, prev_cme_id, prev_cme_diff, n_cmes_before, n_cmes_before_1, n_cmes_before_2, n_cmes_before_3, n_cmes_before_4, n_cmes_before_5, label, cme_diff, verification_level, cme_id)
VALUES
(?, ?, ?, ?, ?, ?, ?, ?, ?, ?, ?, ?, ?, ?, ?, ?, ?, ?, ?)
""", nrow)
# Except foreign key constraint violations
except sqlite3.OperationalError as e:
print(nrow)
raise e
for row in tqdm(rejected_rows):
nrow = list(row[1])
nrow[0] = int(nrow[0])
nrow = tuple(nrow)
try:
cur.execute("""
INSERT INTO GENERAL_DATASET_REJECTED
(harpnum, lead_in_start, lead_in_end, obs_start, obs_end, pred_start, pred_end, reason, message)
VALUES
(?, ?, ?, ?, ?, ?, ?, ?, ?)
""", nrow)
# Except foreign key constraint violations
except sqlite3.OperationalError as e:
raise e
conn.commit()
conn.close()Finally we write all of these results into the database and include some of the original tables into it as well
And we can run the whole thing
if __name__ == "__main__":
create_indices()
create_temp_table()
accepted_rows, rejected_rows = get_all_rows(24 * u.hour, 1 * u.hour, strict=True, table="NO_LIMB_BBOX")
write_into_database(accepted_rows, rejected_rows)Done
Data reduction summary
| Step | Substep | CMEs | SHARP regions |
|---|---|---|---|
| Starting point | 1001 | 3655 | |
| Generating dataset | BBoxes within 70 degrees | 1001 | 3553 |
| Generating dataset | Extract sequences | 498 | 3551 |
